- Investment
- 科学论坛
-
Program
News Message
Intracellular protein editing enables incorporation of noncanonical residues in endogenous proteins
- by wittx 2025-05-31
Editor’s summary
Once proteins are synthesized by the ribosome, their sequence is set and cannot be easily changed through biochemical modifications. Beyer et al. developed a method to switch entire protein segments. In this approach, two split inteins, encoded elements that are self-cleaving when joined, enable the removal and replacement of an interior segment of a protein (see the Perspective by Hampton and Liu). The authors demonstrated that their system functions in vitro and in live cells by installing modified peptide tags into endogenously expressed tagged proteins. —Michael A. FunkStructured Abstract
INTRODUCTION
The study of proteins in situ in mammalian cells is essential to advance our understanding of eukaryotic cell biology. To study proteins in live mammalian cells, an experimental handle is required to track or control a protein of interest throughout experiments. Often, this requires the addition of amino acids—ranging from relatively small peptides to larger fusion proteins—to the target protein. These tags have revolutionized the study of proteins in cells but have limitations: Tags can influence the properties of target proteins and largely lack temporal resolution for studying dynamic processes. Chemical labels benefit from their small size and diversity beyond genetically encodable residues, yet they remain difficult to apply to proteins in mammalian cells. It is clear that new technologies are needed to label and manipulate proteins in live mammalian cells.RATIONALE
Motivated by recent advances in chemical biology and gene editing, we envisioned a technology that could enable the posttranslational editing of protein primary sequences inside mammalian cells, including endogenous proteins that best represent native biology. This system would facilitate splicing of an exogenous protein segment into a target protein at a user-defined site, with temporal control and access to useful labels and noncanonical amino acids. To achieve this goal, we designed a system for intracellular protein editing that relies on two pairs of split intein proteins, which individually ligate polypeptide chains. Used in tandem, these split inteins enable the editing of a donor peptide into an acceptor site situated internally within a target protein. The acceptor sequence is inserted into a target protein—either an endogenous protein through genome editing or an exogenously expressed protein. The corresponding donor protein is generated recombinantly, allowing the incorporation of noncanonical amino acids at scale and chemical labeling, if desired. The donor protein is delivered into live mammalian cells and results in the rapid editing of the donor into the target protein.RESULTS
We first demonstrate a general editing approach, where a generic and minimal donor containing an epitope tag is edited into model proteins calnexin and β-actin. We confirmed the editing of the primary protein sequence using immunoblotting, fluorescence microscopy, and mass spectrometry–based proteomics. Temporal characterization reveals that editing occurs within 10 min of donor protein delivery, which enables experiments on biologically relevant timescales. We demonstrate a sequence-specific mode of editing, where the transcription factor c-Myc and kinase Chk1 are edited nearly tracelessly, enabling time-resolved studies of these dynamic proteins. Further, we show that Chk1 and c-Myc remain functional after editing using proteomic and transcriptomic approaches, which validates our protein editing technology as near-traceless and minimally perturbing.Additionally, genetic code expansion in Escherichia coli was used to encode the noncanonical amino acid and click chemistry handle p-azido-phenylalanine (pAzF) into the donor protein before labeling with small molecules. We then edited small-molecule fluorophores or biotin into endogenous calnexin, enabling microscopy and pull-down studies of the endogenous protein without the use of antibodies.CONCLUSION
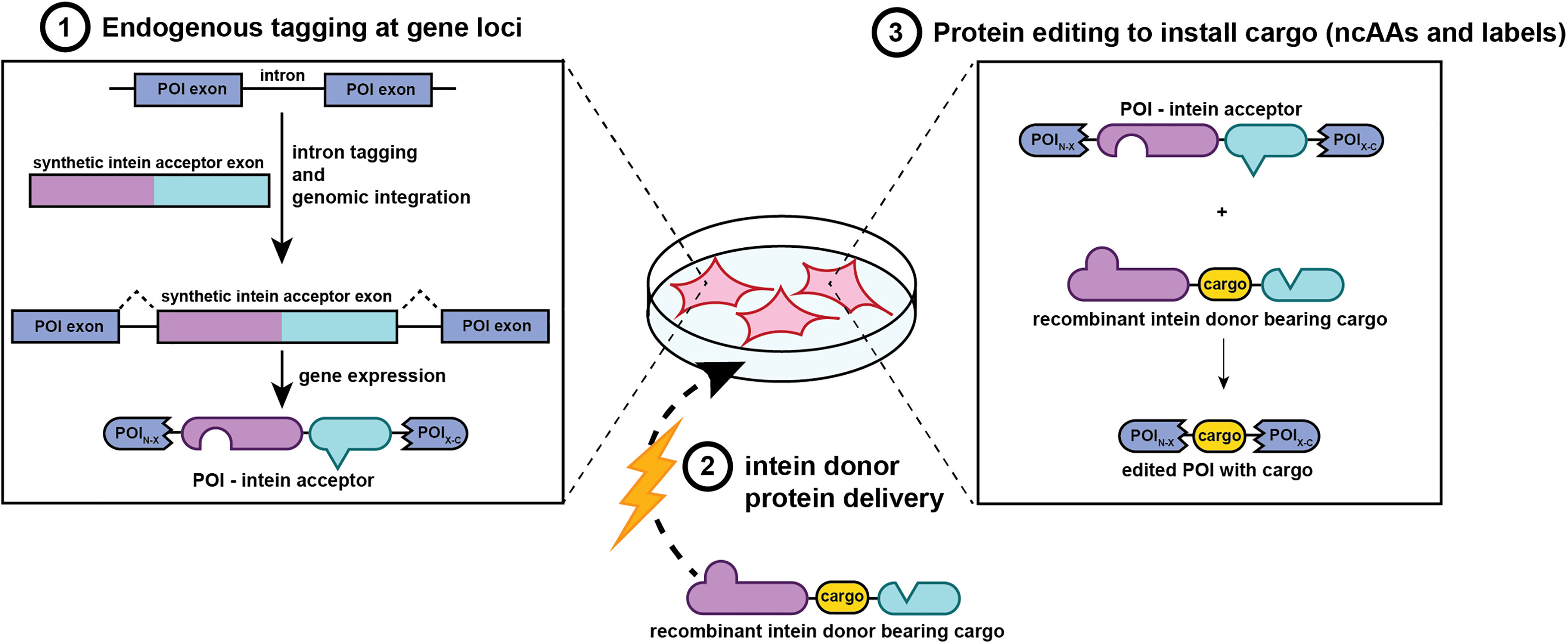
Our work establishes a method for editing the primary amino acid sequence of proteins in live mammalian cells. This approach enables the site-specific editing of noncanonical amino acids and useful labels into target proteins, including endogenous proteins. Ultimately, this enables the labeling and manipulation of proteins in a nondisruptive and temporally resolved fashion. This protein editing method represents a useful and widely applicable addition to the repertoire of technologies for studying proteins in mammalian cells, offering the potential to edit, manipulate, and observe nearly limitless target proteins across diverse applications. Intracellular protein editing.Intracellular protein editing of an endogenous protein of interest (POI) is enabled by a combination of (1) endogenous gene tagging, (2) delivery of a cargo donor protein, and (3) tandem intein trans-splicing to produce a new polypeptide bearing the cargo. ncAAs, noncanonical amino acids.
Intracellular protein editing.Intracellular protein editing of an endogenous protein of interest (POI) is enabled by a combination of (1) endogenous gene tagging, (2) delivery of a cargo donor protein, and (3) tandem intein trans-splicing to produce a new polypeptide bearing the cargo. ncAAs, noncanonical amino acids.Abstract
The ability to study proteins in their native cellular context is crucial to our understanding of biology. In this work, we report a technology for intracellular protein editing, drawing from split intein–mediated protein splicing, genetic code expansion, and endogenous protein tagging. This approach enables us to rapidly and site-specifically install residues and chemical handles into a protein. We demonstrate the power of this platform to edit cellular proteins, inserting epitopes, protein-specific sequences, and noncanonical amino acids. Notably, we use an endogenous tagging approach to apply our protein editing technology to endogenous proteins with minimal perturbation. We anticipate that the protein editing technology presented in this work will be applied to a diverse set of problems and phenomena in live mammalian cells.
Share Http URL: http://www.wittx.cn/get_news_message.do?new_id=1457

Best Last Month